Research
About Our Research
The laboratory uses a novel computational genetic analysis method to identify the genetic factors affecting susceptibility to disease in mouse models. Over 25 genetic factors affecting susceptibility to drug addiction, chronic pain, infectious diseases and others have already been identified. An ongoing effort is now analyzing 8000 biomedical responses in panels of inbred mouse strains. Single cell RNA sequencing and metabolic analysis is used to identify developmental and disease causing pathways. The lab also uses stem cell-based methods for tissue engineering. It has: produced mice with humanized livers, which are used to improve drug safety; developed methods to engineer human liver from adipocyte stem cells; and to produced human liver organoids from iPSC, which are used to study the pathogenesis of human genetic liver diseases.
Computational Genetics and Integrative Biomic Analysis
To reduce the cost and the time frame for genetic research, we have developed a novel computational method that rapidly identifies the genetic basis for biomedical trait differences among inbred mouse strains [1-3] [4].

Conventional (QTL) mouse genetic analysis methods require 5 to 10 years to produce results; while a computational analysis can be completed in less than one day, which significantly accelerates the rate of genetic discovery. In a mapping experiment, a property of interest is measured in >10 inbred mouse strains; genetic factors are then computationally predicted by identifying genomic regions where the pattern of genetic variation correlates with the distribution of trait values among the inbred strains [1]. This innovative approach enables genetic analyses to be completed in far less time (days vs. years), with far fewer personnel, and with a higher overall success rate than could be achieved using conventional mouse genetic analysis methods [3]. Since its inception in 2004, there have been multiple successful demonstrations of its ability to rapidly identify causative genetic factors for biomedical traits in mice, including: gene expression [1]; pharmacogenetic factors [5-8]; susceptibility to invasive Aspergillosis [9] and Respiratory Syncitial Virus infections [10]; analgesic medication [11] and inflammatory [12, 13] and chronic [14] pain; incisional wound biology [15, 16]; haloperidol-induced toxicity [17]; and 4 genes affecting narcotic drug responses [11, 18-20]. The latter genetic discovery generated a novel treatment strategy for narcotic drug withdrawal, which was shown to be effective in control human subjects [20]. There is an ongoing NIH funded 5-center clinical trial that is testing whether this approach can reduce drug withdrawal symptoms in babies born to mothers that consume narcotics.
To attack 21st Century biomedical problems, the lab is applying integrative genomic (genetic, metabolomic, transcriptional and knowledge-based) approaches that will accelerate the rate of translational biomedical discovery. These integrated multi-dimensional studies of biomedical systems will be referred to as ‘Biomic Analysis.’ The level of thousands of mRNAs, metabolites or proteins within a tissue can now be simultaneously measured; which provide orthogonal information to help eliminate false positive results. Our recent analysis of a murine genetic model of acetaminophen (Tylenol)-induced liver toxicity provides an exciting example of how Biomic analysis can lead to exciting discoveries that can provide potential solutions to a major public health problem. Acetaminophen is a safe and effective drug when administered appropriately, but an acute overdose causes sever liver damage. Because of its widespread use, acetaminophen toxicity has become the most frequent cause of acute liver failure in the United States [21]. To uncover the genetic basis for inter-strain differences in susceptibility to this toxicity; we performed an integrative genetic, transcriptional, and metabolomic analysis of the drug-induced response in livers obtained from resistant and sensitive strains. Changes in endogenous metabolites (1H-13C 2-dimensional-NMR) [22] and gene expression (microarrays) in liver were simultaneously examined at 0, 3 and 6 hr after acetaminophen exposure [23]. This integrative analysis identified homocysteine S-methyl transferase 2 (Bhmt2) as a diet-dependent genetic factor that affected susceptibility to acetaminophen-induced liver toxicity in mice. Through an effect on methionine and glutathione biosynthesis, Bhmt2 could utilize its substrate [S-methylmethionine (SMM)] to confer protection against acetaminophen-induced injury in vivo. This work demonstrates how an integrative genomic analysis in mice can provide a unique and clinically applicable approach to a major public health problem.
Mice with ‘Humanized’ Livers

This project was selected by the NIH director for a Transformative R01 Award in October of 2010.
A novel experimental in vivo platform that replaces mouse liver with functioning human liver tissue was developed in collaboration with investigators at the Central Institute for Experimental Animals (Japan). To do this, a thymidine kinase (TK) transgene was expressed within the liver of highly immunodeficient mice (TK-NOG). Mouse liver cells expressing this transgene were ablated after a brief exposure to a non-toxic dose of gancyclovir, and transplanted human liver cells were stably maintained within the liver (humanizedTK-NOG) without exogenous drug or immunosuppressive treatments. The reconstituted liver was shown to be a mature and functioning “human organ.” It had zonal position-specific enzyme expression and bile duct function representative of mature human liver, and could generate a human-specific profile of drug metabolism. These features make the TK-NOG mouse the preferred experimental platform for in vivo analysis of drug metabolism or liver regeneration. The mice are maintained in a specialized barrier facility that was designed for housing these mice. Three areas of interest have emerged from our studies using this model.
1) We recently demonstrated that our chimeric mice could identify drugs that will cause liver toxicity in humans, which was not predicted by toxicology studies in conventional animal species. For example, 7 of 15 subjects treated with a drug (FIAU) in 1992 developed acute liver failure and died. Toxicology studies in mice, rats, dogs and primates did not provide any indication of this drug’s toxicity. However, the clinical features and liver pathology developing in FIAU-treated chimeric TK-NOG mice mirrored those of human subjects [24]. This paper was selected as one of the most influential and impactful papers of the past decade by PLoS editors. Another recent paper demonstrates the utility of the humanized mice for predicting that a drug will cause cholestatic toxicity [25].
2)We developed a method for differentiating iPS cells into induced hepatocytes and demonstrated that they could also reconstitute the human liver. From this work, we received NIH funding (1R01DK102182-01A1 PI: Peltz, G.) to produce in vivo stem cell-based models for 2 human genetic liver diseases.
3) We also developed a novel and patented method for differentiating adipocyte stem cells into induced hepatocytes [26]. This method is orders of magnitude more efficient than existing methods; it is completed in 9 days, and is performed in fully defined media. We have scaled it to process liter volumes of liposuction material, and is now GMP-compliant. We have met with the FDA, and are now completing the pre-clinical studies required for a clinical trial. A multi-disciplinary clinical team has been formed, and we hope to begin a human liver regeneration clinical trial within 12 months.
“Human Liver Organoid”
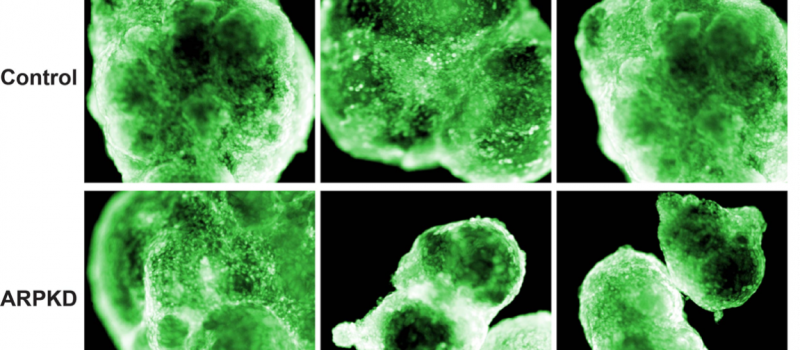
Background information on our hepatic organoid (HO) system. (i) We produced an in vitro system that differentiates a human iPSC into a multi-lineage HO in response to the sequential addition of specific growth factor combinations. It is the only system where a 3-D HO with hepatocytes, cholangiocytes and bile ducts are produced from iPSC. We also developed a CRISPR/PiggyBac system for efficiently engineering disease-causing mutations into the genome of iPSCs and demonstrated that HOs provide a unique system for characterizing the impact of liver-disease-causing genetic mutations (1). (ii) We used scRNA-Seq and metabolomics to analyze the early stages of HO development and identified a biosynthetic pathway that is essential for early liver development. A commonly used over the counter medication (meclizine) was found to inhibit the rate-limiting enzyme in this pathway, and we demonstrated that it could be repurposed for treatment of liver cancer (2). (iii) A more extensive scRNA-Seq analysis of HO cultures at various differentiation stages revealed that HOs also contain cells of other lineages and formed more complex structures (primitive venular structures) than was appreciated in our earlier studies (3). Moreover, we demonstrated that HOs engineered to express the most common causative mutation for Autosomal Recessive Polycystic Kidney Disease (ARPKD) developed the key features of ARPKD liver pathology (abnormal bile ducts, hepatic fibrosis) in only 21 days. Second harmonic generation microscopy confirmed that the ARPKD organoids had a marked increase in thick collagen fibers, and scRNA-Seq analysis was used to characterize the pathways regulating the conversion of hepatic stellate cells into collagen producing myofibroblasts (3).
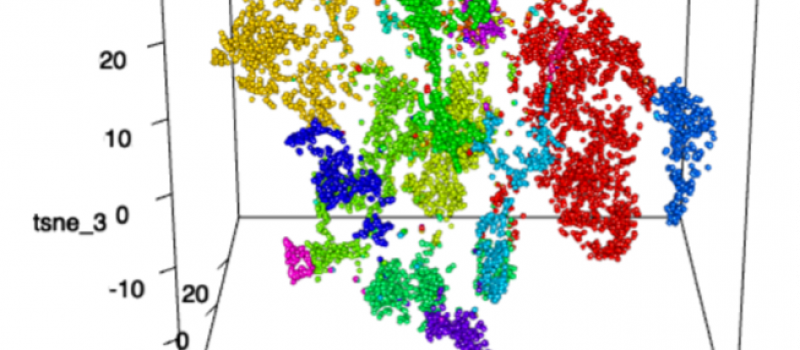
scRNA-seq revealed different clusters between control and ARPKD HOs.
Potential anti-fibrosis drugs. HOs were treated with PDGFRB inhibitor.
References
- Liao G, Wang J, Guo J, Allard J, Cheng J, Ng A, Shafer S, Puech A, McPherson JD, Foernzler D et al: In Silico Genetics: Identification of a Functional Element Regulating H2-Ea Gene Expression. Science 2004, 306(5696):690-695.
- Wang J, Peltz G: Haplotype-Based Computational Genetic Analysis in Mice. In: Computational Genetics and Genomics: New Tools for Understanding Disease. Totowa, New Jersey: Humana Press Inc.; 2005: 51-70.
- Wang J, Liao G, Usuka J, Peltz G: Computational Genetics: From Mouse to Man? Trends in Genetics 2005, 21(9):526-532.
- Zheng M, Dill D, Peltz G: A better prognosis for genetic association studies in mice. Trends in Genetics 2012, 28(2):62-69.
- Guo YY, Weller PF, Farrell E, Cheung P, Fitch B, Clark D, Wu SY, Wang J, Liao G, Zhang Z et al: In Silico Pharmacogenetics: Warfarin Metabolism. Nature Biotechnology 2006, 24:531-536.
- Guo YY, Liu P, Zhang X, Weller PMM, Wang J, Liao G, Zhang Z, Hu J, Allard J, Shafer S et al: In vitro and In silico Pharmacogenetic Analysis in Mice. Proceedings of the National Academy of Sciences 2007, 104:17735-17740.
- Liao G, Zhang X, Clark DJ, Peltz G: A genomic “roadmap” to “better” drugs. Drug Metabolism Reviews 2008, 40(2):225-239.
- Zhang X, Liu H-H, Weller P, Tao W, Wang J, Liao G, Zheng M, Monshouwer M, Peltz G: In Silico and In Vitro Pharmacogenetics: Aldehyde Oxidase Rapidly Metabolizes a p38 Kinase Inhibitor. The Pharmacogenomics Journal 2011, 11(1):15-24.
- Zaas AK, Liao G, Chein J, Usuka J, Weinberg C, Shore D, Giles D, Marr K, Burch L, Perara et al: Plasminogen Alleles Influence Susceptibility to Invasive Aspergillosis. PLoS genetics 2008, 4(6):e1000101.
- Tregoning JS, Yamaguchi Y, Wang B, Mihm D, Harker JA, Bushell ESC, Zheng M, Liao G, Peltz G, Openshaw PJM: Genetic Susceptibility to the Delayed Sequelae of RSV Infection is MHC-Dependent, but Modified by Other Genetic Loci. J Immunology 2010, 185(6):5384-5391.
- Smith SB, Marker CL, Perry C, Liao G, Sotocinal SG, Austin JS, Melmed K, David Clark J, Peltz G, Wickman K et al: Quantitative trait locus and computational mapping identifies Kcnj9 (GIRK3) as a candidate gene affecting analgesia from multiple drug classes. Pharmacogenetics and Genomics 2008, 18(3):231-241.
- LaCroix-Fralish ML, Mo G, Smith SB, Sotocinal SG, Ritchie JG, Austin JS, Melmed K, Schorscher-Petcu A, Laferriere AC, Lee THet al: The β3 Subunit of the Na+,K+-ATPase Affects Pain Sensitivity. Pain 2009, 144:294-302.
- Li X, Sahbaie P, Zheng M, Ritchie J, Peltz G, Mogil JS, Clark JD: Expression genetics identifies spinal mechanisms supporting formalin late phase behaviors. Molecular Pain 2010, 6:11.
- Sorge RE, Trang T, Dorfman R, Smith SB, Beggs S, Ritchie J, Austin J-S, Zaykin DV, Meulen HV, Costigan M et al: Genetically determined P2X7 receptor pore formation regulates variability in chronic pain sensitivity. Nature Medicine 2012, 18(4):595-600.
- Hu Y, Liang D, Li X, Liu H-H, Zhang X, Zheng M, Dill D, Shi X, Qiao Y, Yeomans D et al: The Role of IL-1 in Wound Biology Part I: Murine in Silico and In vitro Experimental Analysis. Anesthesia & Analgesia 2010, 111(6):1525-1533.
- Hu Y, Liang D, Li X, Liu H-H, Zhang X, Zheng M, Dill D, Shi X, Qiao Y, Yeomans D et al: The Role of IL-1 in Wound Biology Part II: In vivo and Human Translational Studies. Anesthesia & Analgesia 2010, 111(6):1534-1542.
- Zheng M, Zhang H, Dill DL, Clark JD, Tu S, Yablonovitch AL, Tan MH, Zhang R, Rujescu D, Wu M et al: The Role of Abcb5 Alleles in Susceptibility to Haloperidol-Induced Toxicity in Mice and Humans PLoS Medicine 2015, 12(1):e1001782.
- Liang D, Liao G, Wang J, Usuka J, Guo YY, Peltz G, Clark JD: A Genetic Analysis of Opioid-Induced Hyperalgesia in Mice Anesthesiology 2006, 104:1054-1062.
- Liang DY, Liao G, Lighthall G, Peltz G, Clark JD: Genetic Variants of the P-Glycoprotein Gene Abcb1b Modulate Opioid-Induced Hyperalgesia, Tolerance and Dependence. Pharmacogenetics and Genomics 2006, 16:825-835.
- Chu LF, Liang D-Y, Li X, Sahbaie P, D’Arcy N, Liao G, Peltz G, Clark JD: From Mouse to Man: The 5-HT3 Receptor Modulates Physical Dependence on Opioid Narcotics. Pharmacogenetics and Genomics 2009, 19:193-205.
- Larson AM, Polson J, Fontana RJ, Davern TJ, Lalani E, Hynan LS, Reisch JS, Schiodt FV, Ostapowicz G, Shakil AO et al: Acetaminophen sets records in the United States: number 1 analgesic and number 1 cause of acute liver failure. Liver Transplantation 2006, 12(4):682-683.
- Zheng M, Lu P, Pease J, Liao G, Peltz G: An Automated Method for Analysis of 2-Dimensional 1H 13C NMR Spectra. Bioinformatics 2007, 23:2926-2933.
- Liu H-H, Lu P, Guo Y, Farrell E, Zhang X, Zheng M, Bosano B, Zhang Z, Allard J, Liao G et al: An Integrative Genomic Analysis Identifies Bhmt2 As A Diet-Dependent Genetic Factor Protecting Against Acetaminophen-Induced Liver Toxicity Genome Research 2010, 20:28-35.
- Xu D, Nishimura T, Nishimura S, Zhang H, Zheng M, Guo Y-Y, Masek M, Michie SA, Glenn J, Peltz G: Fialuridine Induces Acute Liver Failure in Chimeric TK-NOG Mice: A Model for Detecting Hepatic Drug Toxicity Prior to Human Testing PLOS Medicine 2014, 11(4):e1001628.
- Xu D, Wu M, Nishimura S, Nishimura T, Michie SA, Zheng M, Zichengg Y, Yates AJ, Day JS, Hillgren KM et al: Chimeric TK-NOG Mice: A Predictive Model for Cholestatic Human Liver Toxicity. J Pharm Exp Ther 2015, 352:274-280.
- Xu D, Nishimura T, Zheng M, Woo M, Su H, Sato N, Lee G, Michie S, Glenn J, Peltz G: Enabling Autologous Human Liver Regeneration With Differentiated Adipocyte Stem Cells. Cell Transplantation 2014, 23:1573-1584.

